2. Shortcut Removal using Diffusion Models
Prologue
May has been a busy month, as I had to prepare for (and attend) 4 exams, as well as creating curriculum for the introduction to python and numerical algorithms for solving differential equations used in this UNF summer camp, where I am volunteering.
So instead of rushing a project, now that exams are done, I thought it would be interesting to share my exam project from the DTU course, “Advanced Deep Learning in Computer Vision”. The project was done in collaboration with 2 awesome guys, Joakim Dihn and Vebjörn Skre, with the professor Aasa Feragen as supervisor (who also happened to supervise my Bachelor’s thesis)
Project Formulation
This project is about removing shortcuts from lesion images. In classification, a “shortcut” is some kind of proxy in the image, that is highly correlated with a specific class, without being the actual object of interest. An example would be how a model that is supposed to distinguish between a wolf and a husky might look for snow in the background. This is because images of wolves are usually taken in cold places (usually with snow). This becomes a problem when an image of a wolf in either not in a snowy area, or we get an image of a husky that is in a snowy area:

In this project, the dataset consists of images of lesions of 7 classes (disease types + benign), but we are also provided with a label of whether the image contains a known shortcut or not. Shortcuts in this case are usually ruler markings on the skin.
Our goal is then to remove those shortcuts by first noising the image, and then recreate the image with the shortcut gone - esentially creating a counterfactual explanation of what an image would look like without the shortcut there. This is meant as a preprocessing step that can be applied to a dataset, such that the model that classifies lesion diseases will never be impacted by shortcuts, which lets the model focus on relevant features. The methods are based on this paper.
Understanding the Problem
From the very start, we had a hard time really understanding the nuance of the proposed method. They put a lot of emphasis on the use of DDIM instead of DDPM. We thought that the only change here was to remove the variance of the standard DDPM denoising process. We now realize that the paper also proposed a deterministic forward process that let us “encode anatomical information”. The lack of stochasticity is good, since we are not interested in creating a more expressive image. The only source of stochasticity in the denoising process comes from how we guide the noise prediction toward the “non-shortcut” class.
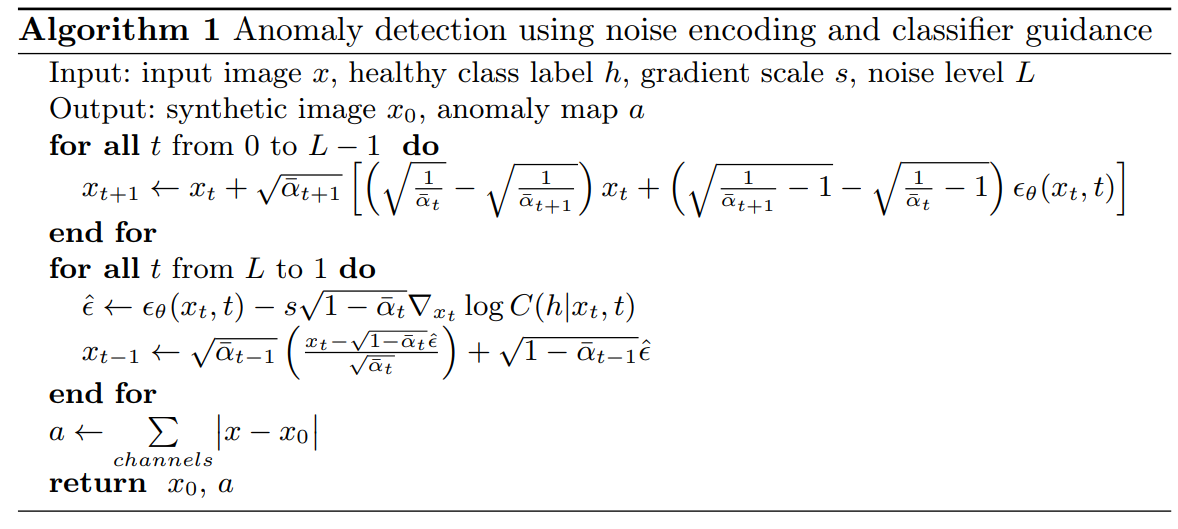
Notice in the forward process how the only “external noise” comes from the noise predictor. This makes it so that if no classifier guidance was present, we could reproduce the original image exactly through the denoising process. Speaking of classifier guidance, it took us some time to realize that using classifier guidance to guide the denoising toward the non-shortcut class was much better than training a diffusion model unconditionally (only training it on images without shortcuts). In both cases, the goal is to guide the denoising toward the distribution of images that don’t include shortcuts. But, with classifier guidance, we can tune the inference by not only using the noise level L, but also the gradient scale s.
Dataset and Artificial Shortcuts
To simplify the task at hand, we reduced the dataset to two classes: “Melanoma diagnosis” and “Non-melanoma diagnosis”. Even though a “healthy” vs. “non-healthy” class would be more desirable; it just wasn’t an option. We wanted to see how much the shortcuts impacted predictions, but as the shortcuts themselves weren’t very clear, we introduced artificial green shortcuts that we would add to many “Melanoma” images. This was also done to increase the correlation between shortcuts and the melanoma class.
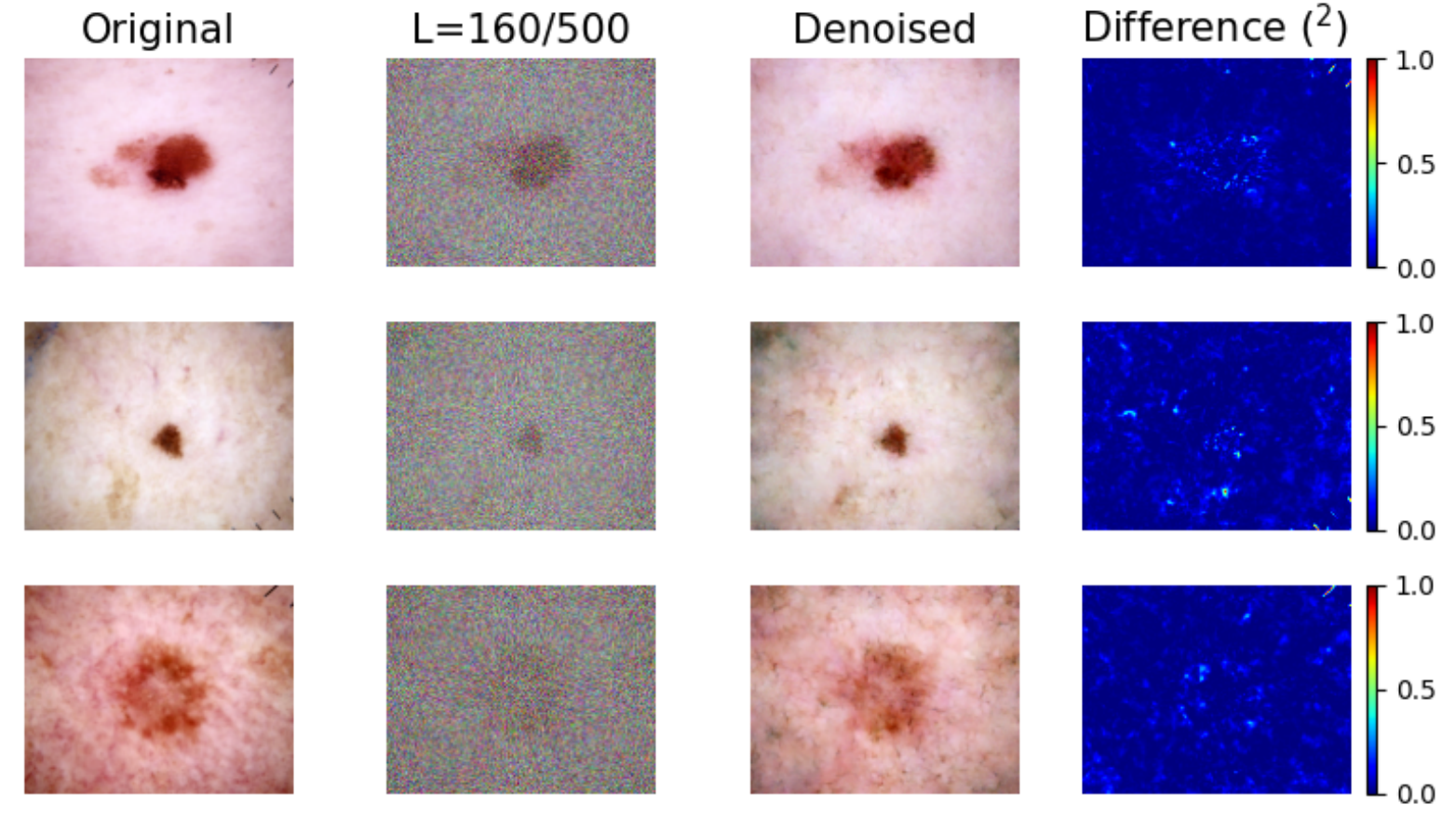
Removal of original shortcuts:
Removal of artificial shortcuts:
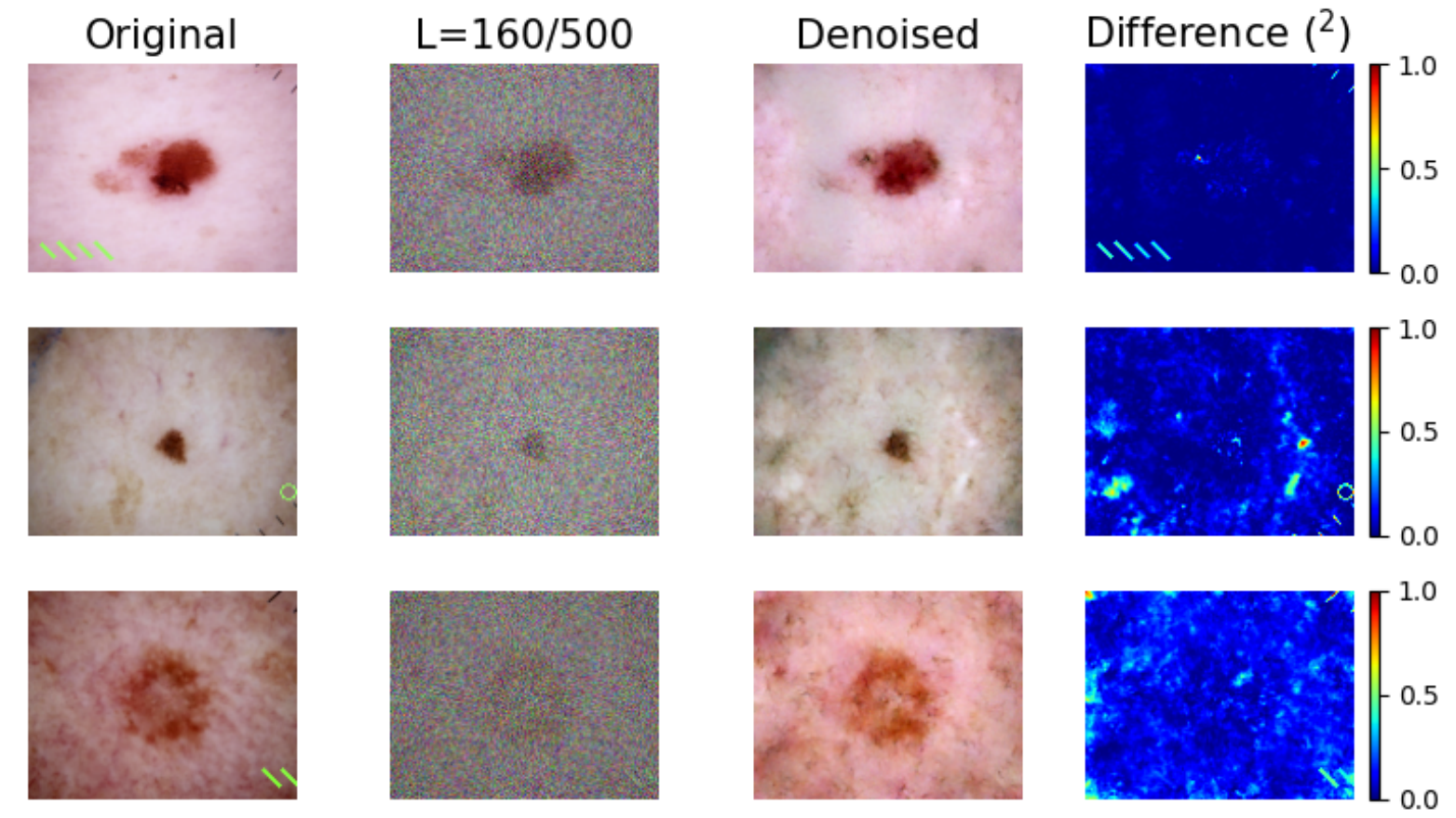
Although not perfect, the impact of removing an artificial shortcut is way more clear.
Classifier Results
To check what a model “sees” when it makes a prediction, we have trained a Visual Transformer model (ViT) that shows which areas have higher associated attention. The model has been trained on the dataset with artificial shortcuts. The following first shows an example of an image from the artificial shortcut dataset, and below it, an image with the shortcut removed through diffusion:
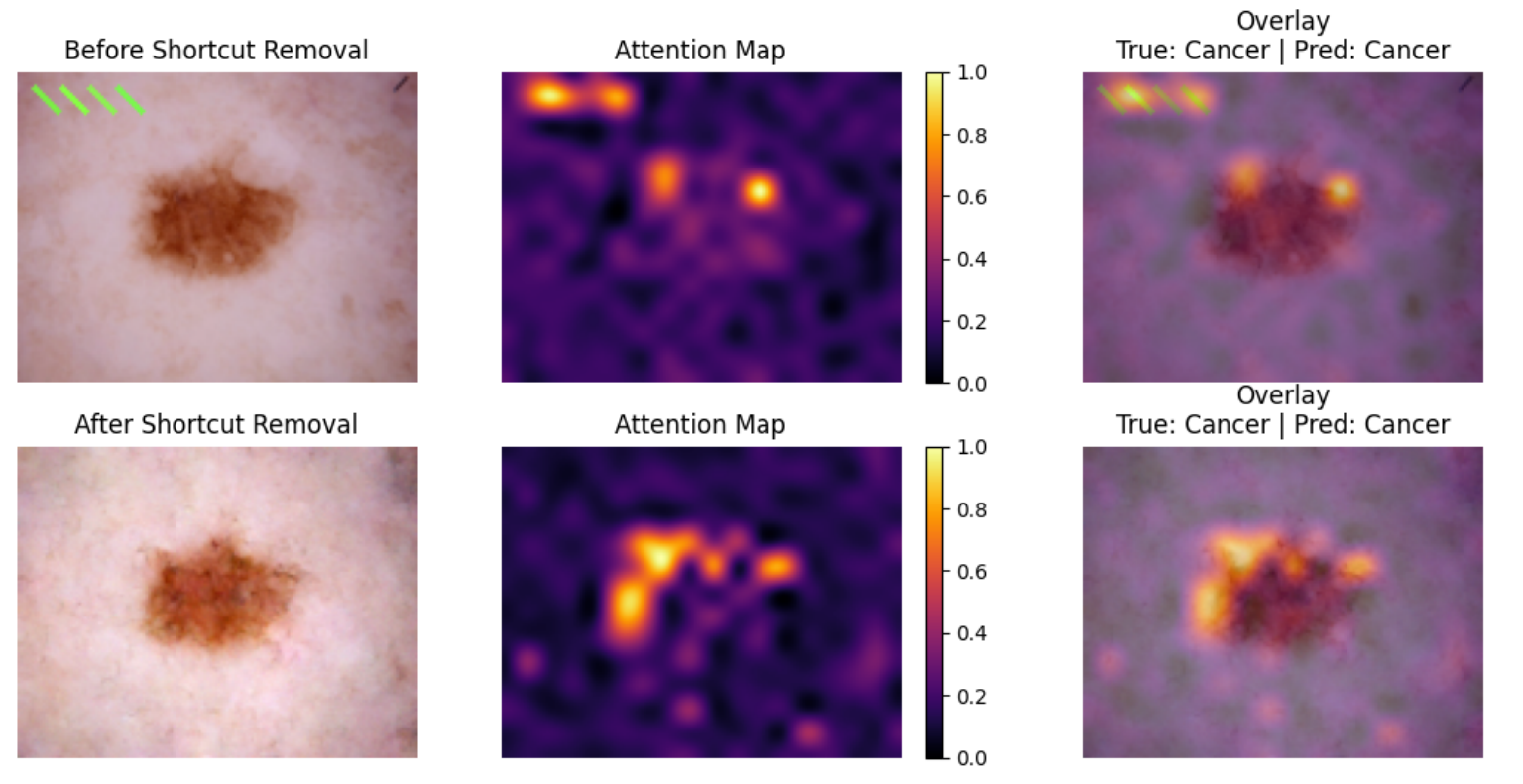
It is very obvious from this, that the model abuses the shortcut when it’s present. When it’s not present, it actually looks at the lesions. More specifically, it look at the edge of the lesion, which is also what professional doctors use when looking for signs of disease.
We have also tried to compare accuracy of models. In the first case, the models have been trained and tested on the artificial shortcut dataset, and in the second one, the models have been trained on the dataset with artificial shortcuts removed - through diffusion, and tested on images with artificial shortcuts:
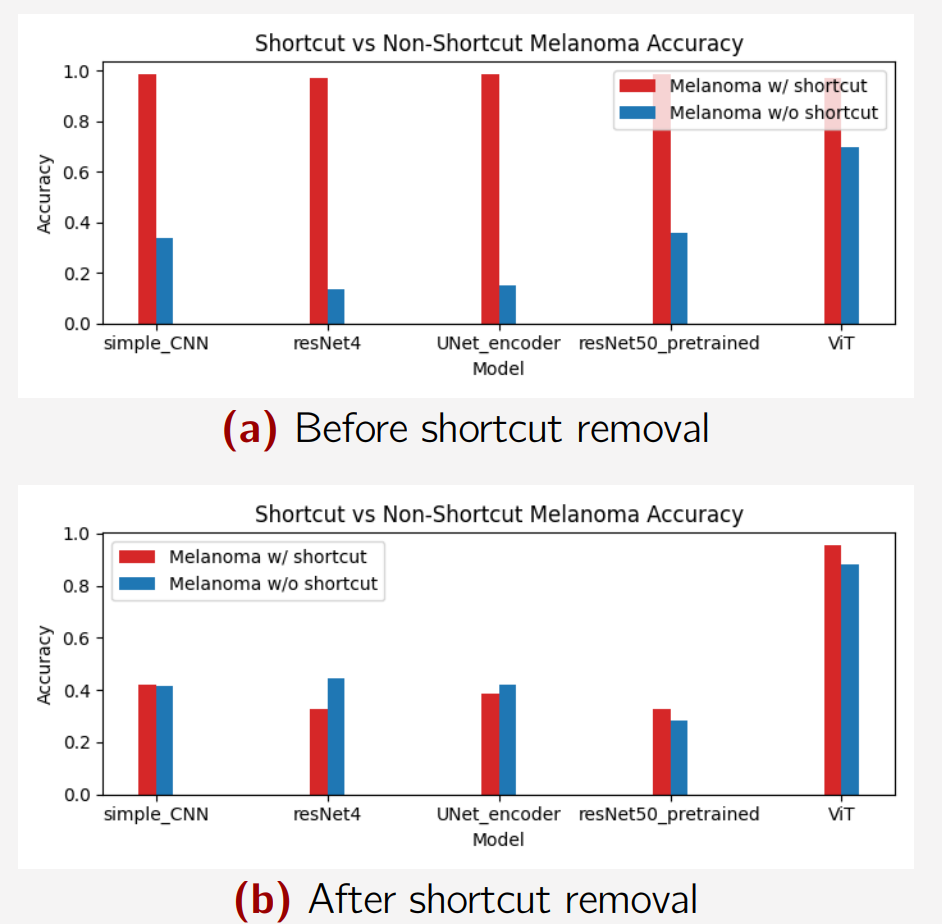
There are two bars, indicating the predictive ability of the model on shortcut- and non-shortcut images, respectively.
The top plot seems to completely over-utilize the shortcuts, as they get almost perfect predictions when shortcuts are present. They also perform very poorly when a shortcut is then not present.
The bottom plot shows the effect of removing shortcuts altogether (but testing on images with shortcuts). The models don’t seems to perform significatly differently despite shortcuts being present. On top of that, the accuracy even increases for the images without shortcuts, as the model can now focus on the actual lesions.
It should be noted that an accuracy below 50% in a binary classification task is a bit weird, since we would then just flip the prediction to get a better accuracy. We are aware of the “bad quality” of the models. The important thing is, that the model has a better understanding of the classes. We do still get a higher accuracy than before.
Discussion and Conclusion
I think the quality of shortcut removal is quite good, but not nearly as impressive as in the paper. As we didn’t utilize the deterministic forward process, our diffusion model altered too much of the images, despite using the correct denoising process. It might have helped to find an even better pair of parameters for the noise level (L=160) and gradient scale (s=100).
Despite that, the still managed to increase generalizability. The lesion classifiers then just had to be trained better, such that it would reach accuracies of >50%